Horizontal Gene Transfer
Current Funding: NSF/EAR Hydrologic Sciences, Collaborative Research: Experiments and Modeling in Horizontal Gene Transfer (co-PI with Helen Nguyen/UIUC and A. Massoudieh/CUA), $129,000 to UC Davis, 2011-2014.
Postdoctoral Associates: Tamir Kamai (now at ARO, Israel).
Students: Arjun Natarajan (Biochemistry undergraduate research assistant), Daniel Nguyen (Physics undergraduate research assistant), Andrew Benjamin.
Collaborators: Helen Nguyen, UIUC; Arash Massoudieh, CUA.
Project Summary: Horizontal gene transfer is widespread among bacteria and is an important component of microbial evolution and trait dispersal. Gene transfer can create new metabolic capabilities, e.g., for degradation of new anthropogenic contaminants, and can facilitate the dissemination of existing capabilities, such as antimicrobial resistance. Soil surfaces provide a natural reservoir for transferrable DNA because sorbed DNA avoids aqueous phase enzymatic degradation and can transform soil bacteria. Cell motility can exert primary control on the frequency with which cells associate with surfaces, and therefore is likely to affect the frequency of interaction between the cells and adsorbed DNA.
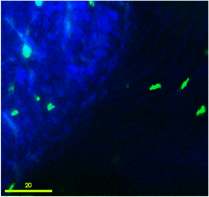
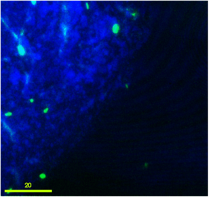
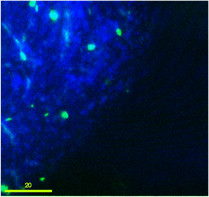
Green-flourescent donor bacterial cells undergoing transport and attachment to sand grain.
This project work investigates the relationships among cell motility, cell attachment and transport, and gene transfer by extracellular DNA. Our goal is to identify controls on the kinetic rate of transformation of bacteria, as a resident-time dependent surface reaction. Transformation should involve truly surface-associated cells, so cells will transform only upon passage through secondary minima to reach primary minima attachment. Thus transformation kinetics of both motile and non-motile cells will depend on their residence-time on surfaces coated with extracellular DNA. The ability of motile cells to swim allows them to approach surfaces independent of aqueous chemistry controls on biocolloid-surface interaction forces.Consequently we expect that motile cells will exhibit greater frequency of transformation in porous media than non-motile cells. We explore this hypothesis through specific experimental and modeling objectives involving determination of attachment mechanisms in a radial stagnation point flow (RSPF) system, of residence time distributions and spatial distribution in a micromodel system, and of attachment-detachment and gene transfer kinetics in batch and column systems. The experiments involve both motile and non-motile bacterial strains and involve surfaces coated with extracellular DNA. Results are used in the development and testing of models of bacterial transport and horizontal gene transfer in porous media.
Important papers on the topic:
- Lu, N., A. Massoudieh, X. Liang, T. Kamai, T. R. Ginn, J. L. Zilles, T. H. Nguyen, Swimming motility reduces Azotobacter vinelandii deposition to silica surfaces, submitted to Environmental Science & Technology, June 2014.
- Massoudieh A., N. Lu, X. Liang, T. H. Nguyen, T. R. Ginn, Bayesian process-identification in bacteria transport in porous media, J. Contam. Hydrol., 153, 78-91, 2013.
- Massoudieh, A., C. Crain, E. Lambertini, K. E. Nelson, T. Barkouki, P. L’Amoreaux, F. J. Loge, T. R. Ginn, Kinetics of conjugative gene transfer among bacteria attached to surfaces in granular porous media, J Contam. Hydrol.,112(1-4): 91-102, 2010.
- Massoudieh A., A. Mathew, E. Lambertini, K. E. Nelson, T. R. Ginn, Horizontal gene transfer on surfaces in natural porous media: Conjugation and kinetics, Vadose Zone Journal 6(2): 306-315, 2007.